Research
I am involved in research activities dealing with quantum chemistry, machine/deep learning, computational drug design and high-performance computing to push pharmacology to the next generation.
Software for Highly-Scalable Biochemistry
I am involved in the MiMiC project together with Prof. Dr. Paolo Carloni, Dr. Jógvan Magnus Haugaard Olsen and Prof. Ursula Röthilsberger. The MiMiC framework is an open-source, general platform that enables the implementation of multiscale computational (bio)chemistry simulation methods through coupling of multiple external programs, designed for massively parallel applications with a multiple-programs multiple-data model. The program is written in a combination of Fortran and C++, and parallelizaed with a hybrid MPI/OpenMP approach. Currently, MiMiC couples the CPMD and GROMACS codes to perform highly scalable QM/MM biomolecular simulations on HPC systems.
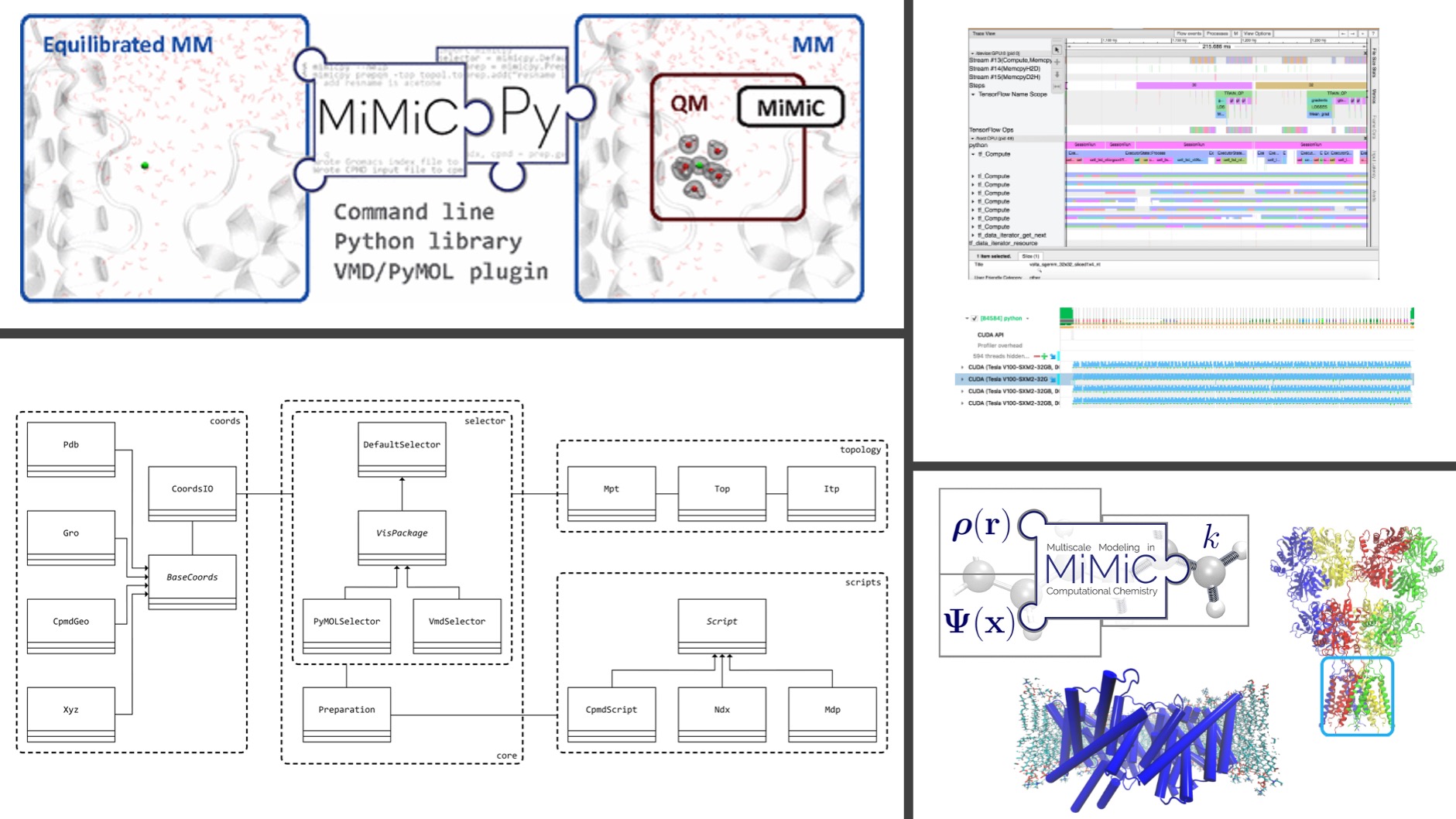
I am the lead developer of MiMiCPy, an open-source Python toolkit as a companion to MiMiC. The code greatly simplifies the preparation and debugging of MiMiC-based QM/MM input files via a user-friendly interface. It provides an extensive list of command-line tools with an easy-to-use selection language to pick out the QM region. Plugins for PyMOL and VMD alow the selection of complex QM regions visually. MiMiCPy can also be used as a Python library, allowing one to develop complex workflows to set up MiMiC-based QM/MM simulations. The package has been designed with a modular and object-oriented approach. This allows one (i) to easily support new topology and coordinate file formats from different programs, when they become available in MiMiC; (ii) to develop new tools as MiMiC expands its functionalities.
I was involved in early efforts to couple MiMiC-QM/MM simulations with machine learning-based force fields. This was carried out as part of the Helmholtz GPU Hackathon 2020 in collaboration with Andrea Rizzi, Florian Schakerth and Viacheslav Bolynkh. I designed a MiMiC data reader for the PiNN library, and contributed to parallelization efforts of the tensorflow-based atomic neural network on multiple GPUs. I also contribute to testing, documentation and communication activities related to MiMiC. I spearheaded training activities at the CECAM Flagship School on MiMiC held at the École Polytechnique Fédérale de Lausanne (EPFL) on 18 to 22 July 2022 and the MiMiC workshop at the Simulate with EBRAINS event held by the Human Brian Project on 10th Nov 2022.
I am the lead developer of MiMiCPy, an open-source Python toolkit as a companion to MiMiC. The code greatly simplifies the preparation and debugging of MiMiC-based QM/MM input files via a user-friendly interface. It provides an extensive list of command-line tools with an easy-to-use selection language to pick out the QM region. Plugins for PyMOL and VMD alow the selection of complex QM regions visually. MiMiCPy can also be used as a Python library, allowing one to develop complex workflows to set up MiMiC-based QM/MM simulations. The package has been designed with a modular and object-oriented approach. This allows one (i) to easily support new topology and coordinate file formats from different programs, when they become available in MiMiC; (ii) to develop new tools as MiMiC expands its functionalities.
I was involved in early efforts to couple MiMiC-QM/MM simulations with machine learning-based force fields. This was carried out as part of the Helmholtz GPU Hackathon 2020 in collaboration with Andrea Rizzi, Florian Schakerth and Viacheslav Bolynkh. I designed a MiMiC data reader for the PiNN library, and contributed to parallelization efforts of the tensorflow-based atomic neural network on multiple GPUs. I also contribute to testing, documentation and communication activities related to MiMiC. I spearheaded training activities at the CECAM Flagship School on MiMiC held at the École Polytechnique Fédérale de Lausanne (EPFL) on 18 to 22 July 2022 and the MiMiC workshop at the Simulate with EBRAINS event held by the Human Brian Project on 10th Nov 2022.
High-Performance Computing & Quantum Chemistry Against Glioma

I am involved in the Helmholtz European Partnering Project between Forschungszentrum Juelich and Italian Institute of Technology, directly working with Prof. Dr. Paolo Carloni and Dr. Marco De Vivo, and involving Prof. Michele Parrinello, Prof. Guilia Rossetti, and Prof. Bernd Neumaier. The main goal is to apply high performance computing approaches to tackle neurological diseases. I am specifically involved in applying scalable Quantum Mechanics/Molecular Mechanics (QM/MM)-based dynamics to study the IDH1 enzyme catalysis. Variants of IDH1 are involved in glioma and glioblastoma. QM/MM simulations of IDH1 are hoped to lead to an understanding of the catalytic mechanism, and an accurate map of the transition state. This information can be used to suggest transition state analogs, which may be a million times more effective compared to current glioma inhibitors. This will be especially useful in suggesting radiotracers for non-invasive detection of early-stage glioma, an avenue being investigated with our experimental collaborator Prof. Bernd Neumaier.
We apply the MiMiC-QM/MM interface (developed within the project mentioned above) to study the dynamics of the IDH1 enzyme. This is the first application of MiMiC to a large, biologically-relavant enzyme. We have showed that system scales efficiently up to an incredible of almost 40,000 cores with Density Function Theory-B3LYP dynamics. This level of scaling has not been reached before for biological simulations at ab-initio level of theory. We hope that this could serve as strong proof of the potential of highly scalable QM/MM to uncover enzyme function, and to design enzyme inhibitors. We are currently working on the simulation of the mutant-IDH1 catalysis, to understand the disease-causing pathway and design transition state analogs against this.
We apply the MiMiC-QM/MM interface (developed within the project mentioned above) to study the dynamics of the IDH1 enzyme. This is the first application of MiMiC to a large, biologically-relavant enzyme. We have showed that system scales efficiently up to an incredible of almost 40,000 cores with Density Function Theory-B3LYP dynamics. This level of scaling has not been reached before for biological simulations at ab-initio level of theory. We hope that this could serve as strong proof of the potential of highly scalable QM/MM to uncover enzyme function, and to design enzyme inhibitors. We are currently working on the simulation of the mutant-IDH1 catalysis, to understand the disease-causing pathway and design transition state analogs against this.
